Driving Leadership in Gene-Disease Validity
We invest in the expertise needed for ongoing evaluation of new and existing gene-disease associations to drive better variant classification, smart product design, and unique service offerings.
Dedicated Experts
With over 100 years of combined experience in the field of Medical Genetics, the Ambry Genetics Gene Team is a dedicated group of scientists committed to enhancing patient care and treatment outcomes through precise genetic insights.
Evaluation of Evidence
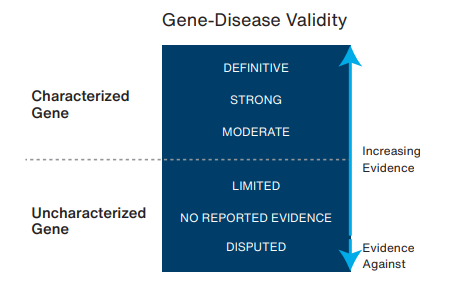
Unique in our field, we have developed a comprehensive gene-disease validity assessment framework, rigorously validated and published in peer-reviewed medical literature, to methodically evaluate the wealth of available genetic and experimental evidence. This not only sets us apart but also ensures that our genetic testing services are at the cutting edge of accuracy and reliability. It also significantly enhances our ability to aid in accurate diagnosis and informed treatment planning as well as to empower patients and their families with valuable knowledge about their genetic health, paving the way for personalized care and improved health outcomes.
Literature Surveillance
As pioneers in gene curation, our core mission is to tirelessly review and analyze the latest scientific evidence, unlocking the complexities of the human genome—one gene at a time. This rigorous approach allows us to deliver the most pertinent genetic insights to our patients with unmatched speed. By doing so, we aim not just to inform but to transform patient care, providing timely, precise, and actionable genetic information that can significantly influence diagnosis, treatment, and overall health management.
Learn More About Gene-Disease Validity Assessments in Hereditary Cancer Testing
Download White Papers and Case Studies Download White Papers and Case StudiesThe Only Commercial Lab with a Peer-Reviewed, Published Gene-Disease Validity Scheme
Improving Variant Classification
With a deep dive into the functions and implications of all 20,000 genes, our experts strive to ensure the utmost accuracy in variant classification. Our deep understanding of gene-disease relationships and firm confidence in the supporting evidence are fundamental in accurately classifying genetic variants and providing meaningful results to our patients. Understanding the true impact of genetic variants on health allows for informed decision-making, personalized treatment strategies and, ultimately, better patient outcomes.
Driving Intelligent Product Design
Our commitment to enhancing patient care through precise genetic insights drives everything we do. If there's uncertainty about whether a gene is linked to a specific condition, then, by default, the pathogenicity—or disease-causing potential—of any variant within that gene remains unclear. At Ambry Genetics, we prioritize establishing clear connections between genes and diseases. By rigorously evaluating gene-disease associations, we ensure that the variants we identify can be accurately classified in terms of their clinical significance.
Enhancing Service to Provide More Answers
We are dedicated to delivering answers to every patient, regardless of when their journey with us began. Our Patient for Life program exemplifies this commitment. Through continuous and meticulous gene curation, we keep our gene-disease database current, ensuring that our insights remain relevant not only for present and future patients but also for those who have previously undergone testing with us.
Videos
Webinars
Peer-Reviewed Papers
- Smith ED, Radtke K, Rossi M, Shinde DN, Darabi S, El-Khechen D, Powis Z, Helbig K, Waller K, Grange DK2, Tang S, Farwell Hagman KD. Classification of Genes: Standardized Clinical Validity Assessment of Gene-Disease Associations Aids Diagnostic Exome Analysis and Reclassifications.Hum Mutat 2017 Jan 20
- Kelly D. Farwell Hagman MS, CGC,Deepali N. Shinde PhD, Cameron Mroske MS, Erica Smith PhD, Kelly Radtke PhD, Layla Shahmirzadi MS, CGC, Dima El-Khechen MS, CGC, Zöe Powis MS, CGC, Elizabeth C. Chao MD, FACMG, Wendy A. Alcaraz PhD, DABMG, Katherine L. Helbig MS, CGC, Samin A. Sajan PhD, Mari Rossi MS, PhL, Hsiao-Mei Lu PhD, Robert Huether PhD, Shuwei Li PhD, Sitao Wu PhD, Mark E. Nuñes MD & Sha Tang PhD, FACMG. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases.Genetics in Medicine, 2016.

