Candidate Disease Gene Discoveries
Clinical Diagnostic Exome sequencing (DES) serves a dual role as a diagnostic and a discovery tool by:
- making large-scale genetic testing more affordable;
- accelerating the rate of scientific discovery of candidate disease genes, thereby increasing our ability to diagnose patients
Clinical reporting of candidate genes identified using DES is based on Ambry’s comprehensive, rule-based scoring criteria. The criteria incorporate Ambry’s gene and variant classification and heavily weigh evidence from studies of gene function, tissue and developmental stage expression, in vitro or in vivo functional studies, knowledge of the gene family and/or pathway, animal models, and familial co- segregation analysis.
Fig. 1 Reports with candidate-gene findings, which are always interpreted as uncertain findings, fall under one of two categories: “candidate” or “suspected candidate” depending upon the level of evidence achieved
(Fig. 1) Candidate Gene Criteria: Scheme to evaluate the level of available evidence to propose a candidate gene-disease relationship
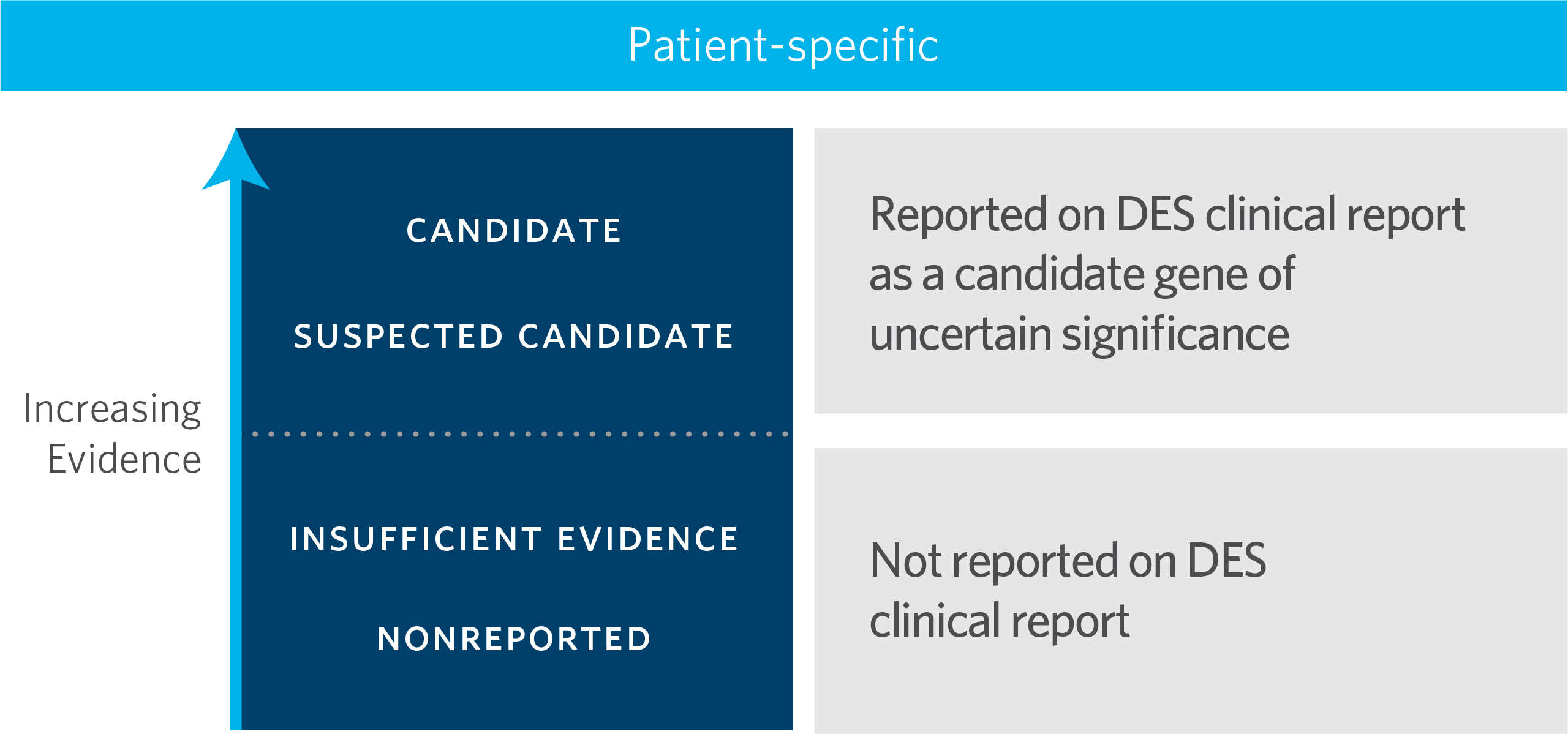
By applying our Candidate Gene Criteria:
We achieve a candidate gene detection rate of 8% among patients referred for diagnostic exome sequencing (Table 1). Among the first 1,500 patients referred for exome sequencing, both characterized and uncharacterized genes were analyzed for 934 patients, among whom 72 (7.7%) had a reported candidate (Table 1). Among these 72 patients, 37 (4.0%) received “candidate” evidence scores and 35 (3.7%) received “suspected candidate” scores.
(Table 1) Candidate Gene Rate Among Those Having ExomeNext, By Evidence Strength
| Evidence Category | Patients With Candidate Gene Results |
|---|---|
| Candidate | 4.0% |
| Suspected Candidate | 3.7% |
| Total | 7.7% |
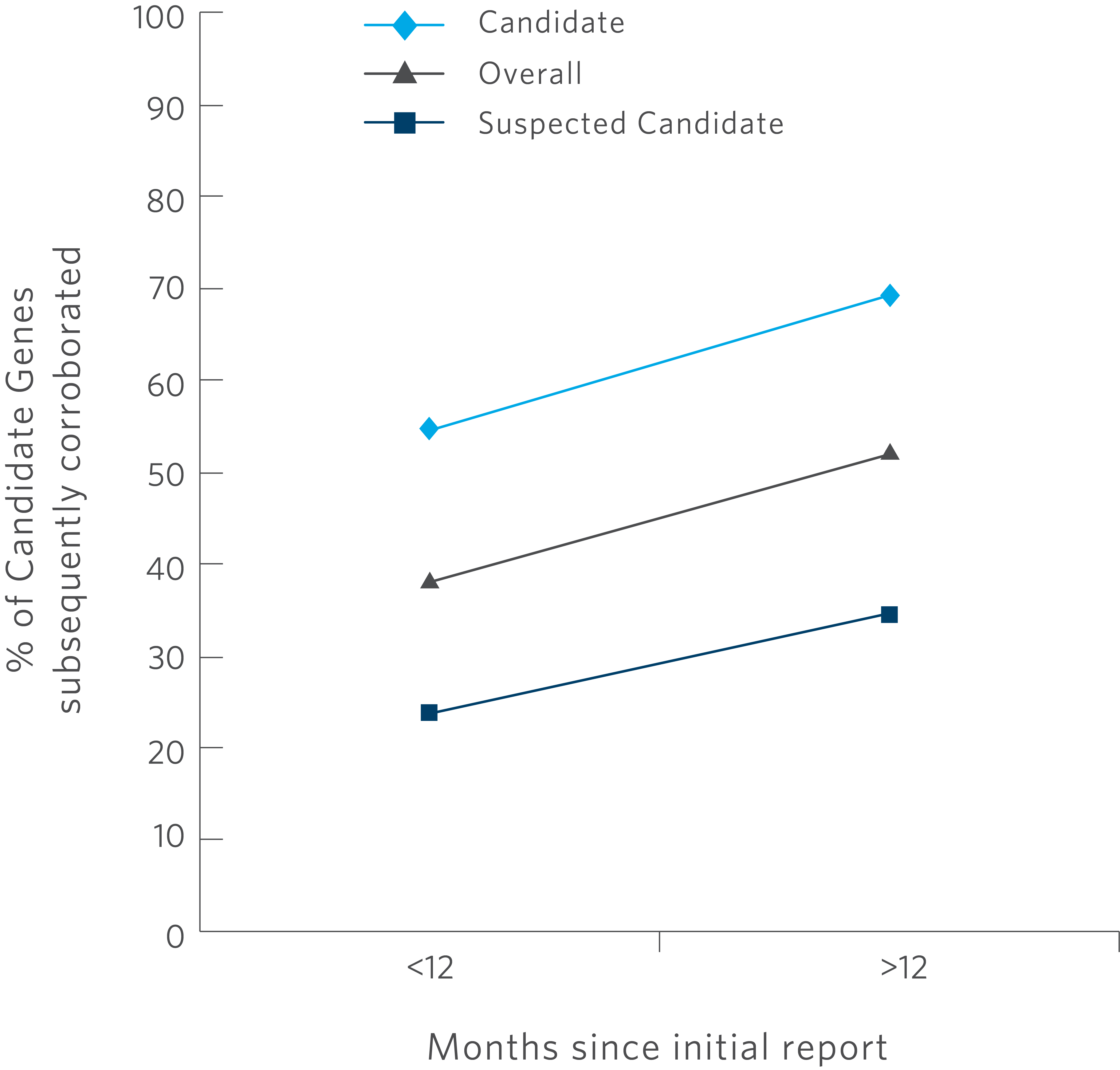
(Fig. 2) Amount of Peer-Reviewed Published Candidate Gene Corroboration Over Time
Over half of all reported candidate genes were subsequently corroborated in the peer-reviewed literature among genes reported >12 months prior (Fig 2). Among candidate genes reaching “candidate” and “suspected candidate” evidence levels, 54.5% (18/33) and 23.7% (9/38) had subsequent corroborating publications, respectively (P = 0.014). The candidate genes described include those reported through November 2015. To determine whether time to publication was a factor for peer-reviewed corroboration, we further restricted the analysis to candidate genes reported through December 2014. In this cohort with at least 12 months available for subsequent corroboration (n = 52), the corroboration rate increased to 51.9% (27/52) overall and to 69.2% (18/26) among genes reaching candidate evidence and 34.6% (9/26) for genes reaching suspected candidate evidence.
Terminology
| Characterized Mendelian disease gene | A gene known to underlie at least one Mendelian genetic condition |
| Uncharacterized Mendelian disease gene | A gene that is not currently known to underlie a Mendelian genetic condition |
| Clinical Validity | Based on existing literature and knowledge about gene-disease relationships, clinical validity is the determination that a particular disease is truly caused by pathogenic variants in a particular gene. |
| Uncharacterized gene-disease relationship | A gene-disease relationship and/or mechanism not previously proposed or with limited evidence based on clinical validity assessment |
| Characterized gene-disease relationship | A disease or phenotype whose underlying molecular etiology is established with at least moderate level of evidence based on ClinGen clinical validity assessment |
| Candidate Gene Criteria | Standardized criteria scheme to evaluate the level of available evidence to propose a candidate gene-disease relationship in a previously uncharacterized gene-disease relationship |
| Idiopathic disease | A disease or phenotype whose underlying molecular etiology or etiologies have not been established. For heterogeneous conditions, there may be multiple etiologies. |
References
1. Kelly D. Farwell Hagman MS, CGC, Deepali N. Shinde PhD, Cameron Mroske MS, Erica Smith PhD, Kelly Radtke PhD, Layla Shahmirzadi, MS, CGC, Dima El-Khechen MS, CGC, Zöe Powis MS, CGC, Elizabeth C. Chao MD, FACMG, Wendy A. Alcaraz PhD, DABMG, Katherine, L. Helbig MS, CGC, Samin A. Sajan PhD, Mari Rossi MS, PhL, Hsiao-Mei Lu PhD, Robert Huether PhD, Shuwei Li PhD, Sitao Wu PhD, Mark E. Nuñes MD & Sha Tang PhD, FACMG. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genetics in Medicine, 2016.
2. Smith ED, Radtke K, Rossi M, Shinde DN, Darabi S, El-Khechen D, Powis Z, Helbig K, Waller K, Grange DK2, Tang S, Farwell Hagman KD. Classification of Genes: Standardized Clinical Validity Assessment of Gene-Disease Associations Aids Diagnostic Exome Analysis and Reclassifications. Hum Mutat, 2017.