
Expanded Coverage with up to 90 Genes
Supporting Patients with Expanded RNA Gene Coverage
Expanded RNA Analysis for Better Variant Classification
+RNAinsight provides comprehensive gene coverage for RNA analysis to help classify DNA variants associated with cancers of breast, ovarian, prostate, colon, pancreatic, uterine, and more. It can be paired with Ambry Genetics hereditary cancer panels to provide functional RNA information to help identify and interpret DNA variants, including deep intronic variants not detected by a DNA-only approach.
+RNAinsight: Paired DNA and RNA Genetic Testing
+RNAinsight improves the sensitivity and clarity of genetic testing. It works in tandem with Ambry Genetics' DNA testing to identify patients with or at risk for hereditary cancer who might otherwise be missed, decrease variants of unknown significance in real-time, and provide more accurate results to inform patient care.
DNA-Only Testing May Miss Patients At Risk For or With Hereditary Cancer
Many patients suspected to have hereditary cancer receive negative or inconclusive results due to limitations of DNA-only genetic testing. While there have been several advancements in genetic testing since the days of Sanger sequencing, there is still a need to enhance variant detection and interpretation. Adding simultaneous RNA genetic testing to DNA-only tests with +RNAinsight is the next step to providing more accurate, actionable results to patients and their families.
Genetic Testing Technology Timeline – DNA and RNA:

Paired DNA and RNA Testing Improves Variant Detection and Classification
Add RNA genetic testing to a hereditary cancer panel for every patient undergoing DNA testing to deliver more clinically actionable results.
+RNAinsight Provides an Additional Line of Evidence To:
Identify More Patients with Hereditary Cancer
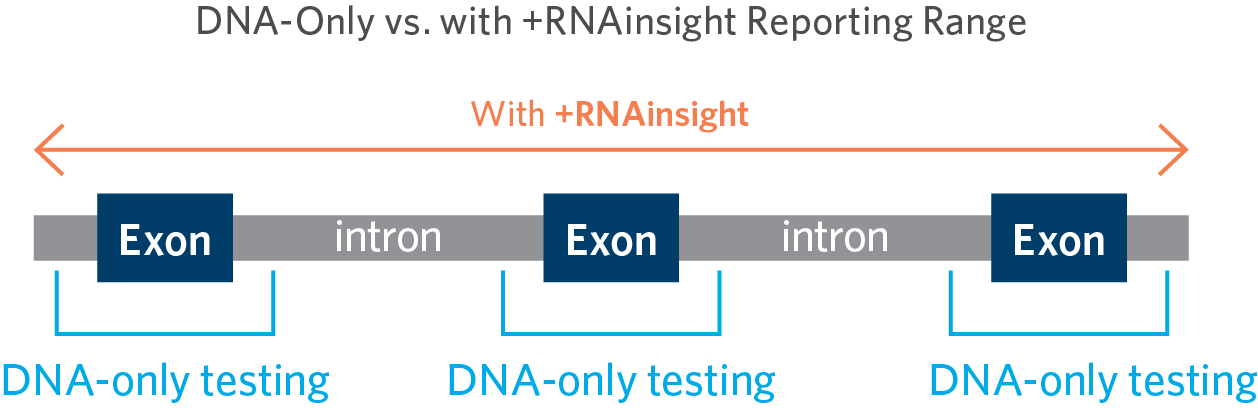
Expanded RNA Analysis for Better Variant Classification
Clinical diagnostic labs typically apply a reporting range limit to their DNA genetic testing panels. Variants outside these ranges have a much lower probability of being pathogenic (disease-causing) and would most often be classified as a variant of unknown significance (VUS). Looking beyond these ranges would only increase the VUS rate without increasing the diagnostic yield. However, using +RNAinsight expands the reporting range of DNA-based testing, which in turn enables Ambry to provide a clear diagnosis to more patients who may otherwise be missed.
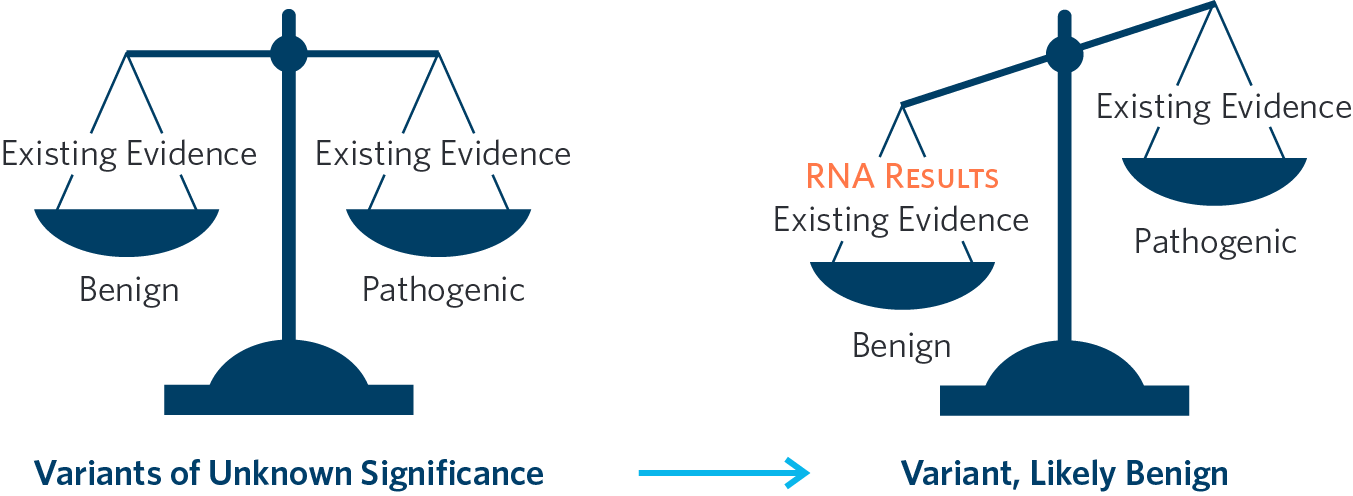
Decrease Variants of Unknown Significance in Real Time
Patients undergoing hereditary cancer testing may receive a VUS result, which can cause uncertainty and confusion for both the ordering healthcare providers and patients. A VUS result does not provide actionable information to inform medical management since it doesn't clarify if the patient is at an increased risk for cancer. +RNAinsight provides an additional line of functional evidence that can tip the scale towards pathogenic or benign. In some cases, this additional evidence can turn a VUS into an actionable positive or clear, negative result.
Provide More Accurate Results to Inform Patient Care
A negative report or a VUS can be unsettling, especially in the context of a strong cancer history. Patients may worry that a mutation was missed by current technology, or that there may be a mutation in another gene. A VUS could be benign, or it could be pathogenic. +RNAinsight analyzes functional RNA data to help classify DNA variants, and as a result, patients receive more sensitive and specific testing, which leads to lowered rates of inconclusive outcomes.
RNA Genetic Testing Decreases Variants of Unknown Significance

Testimonials
+RNAinsight Product Overview
Available to order with hereditary cancer panels
Up to 90 Genes Analyzed by +RNAinsight
AIP ALK APC ATM ATRIP AXIN2 BAP1 BARD1 BMPR1A BRCA1 BRCA2 BRIP1 CDC73 CDH1 CDK4 CDKN1B CDKN2A CEBPA CFTR CHEK2 CPA1 CTNNA1 CTRC DDX41 DICER1 EGFR EGLN1 EPCAM ETV6 FH FLCN GATA2 GREM1 HOXB13 KIF1B KIT LZTR1 MAX MBD4 MEN1 MET MITF MLH1 MLH3 MSH2 MSH3 MSH6 MUTYH NF1 NF2 NTHL1 PALB2 PALLD PDGFRA PHOX2B PMS2 POLD1 POLE POT1 PRKAR1A PRSS1 PTCH1 PTEN RAD51B RAD51C RAD51D RB1 RET RNF43 RPS20 RUNX1 SDHA SDHAF2 SDHB SDHC SDHD SMAD4 SMARCA4 SMARCB1 SMARCE1 SPINK1 STK11 SUFU TERT TMEM127 TP53 TSC1 TSC2 VHL WT1 Expand
14-21 day turnaround time (No increase due to +RNAinsight)
Sample Requirements:
One kit that includes 1 EDTA tube (DNA) and 1 PAXgene® tube (RNA)
** Orders including +RNAinsight are not eligible for STAT testing.
Technical Details
Ribonucleic acid (RNA) is isolated from the patient's specimen using standardized methodology and quantified. RNA is converted to complementary DNA (cDNA) by reverse transcriptase polymerase chain reaction (RT-PCR). Sequence enrichment of the targeted coding exons and adjacent intronic nucleotides is carried out by a bait-capture methodology using long biotinylated oligonucleotide probes followed by polymerase chain reaction (PCR) and Next-Generation sequencing. +RNAinsight analyzes transcripts for up to 90 genes depending which Ambry Genetics DNA based Hereditary Cancer Panel it is paired with and depending on the absence or presence of RNA transcripts expressed in the blood. Any transcripts found are compared to a human reference pool. The absence or presence of RNA transcripts meeting quality thresholds is incorporated as evidence towards assessment and classification of DNA variants. Any regions not meeting RNA quality thresholds are excluded from analysis. The results from +RNAinsight are used to provide functional RNA information to further support classification of DNA variants. It is not intended to be used as a stand-alone diagnostic test.
+RNAinsight: Learn More about the Benefits of DNA and RNA Genetic Testing
References
1. Ambry Genetics, internal data on file2. Karam R. et al. RNA Genetic Testing in Hereditary Cancer Improves Variant Classification and Patient Management. ACMG 2019.
3. Richards S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405-24

