Whole Transcriptome Sequencing and Targeted RNA-seq
Ambry offers both whole transcriptome and targeted RNA analysis. Whole transcriptome studies that analyze differential gene expression patterns can identify signatures that can serve as biomarkers. Gene expression-based biomarkers can define pharmacogenomic associations and serve as companion diagnostics. RNA analysis may also identify novel splice variants and gene fusion transcripts.
Please note that the whole transcriptome analysis service is only available to our biopharma and pharma partners at this time.
Whole Transcriptome Analysis
Ambry uses strand specific sample preparation, which increases confidence in transcript annotation, increases the percentage of aligned reads and reduces overall sequencing cost per sample. We can perform either paired-end sequencing for de novo transcript discovery and isoform detection or single-end sequencing for standard gene expression analysis. ERCC controls are included in all transcriptome projects.
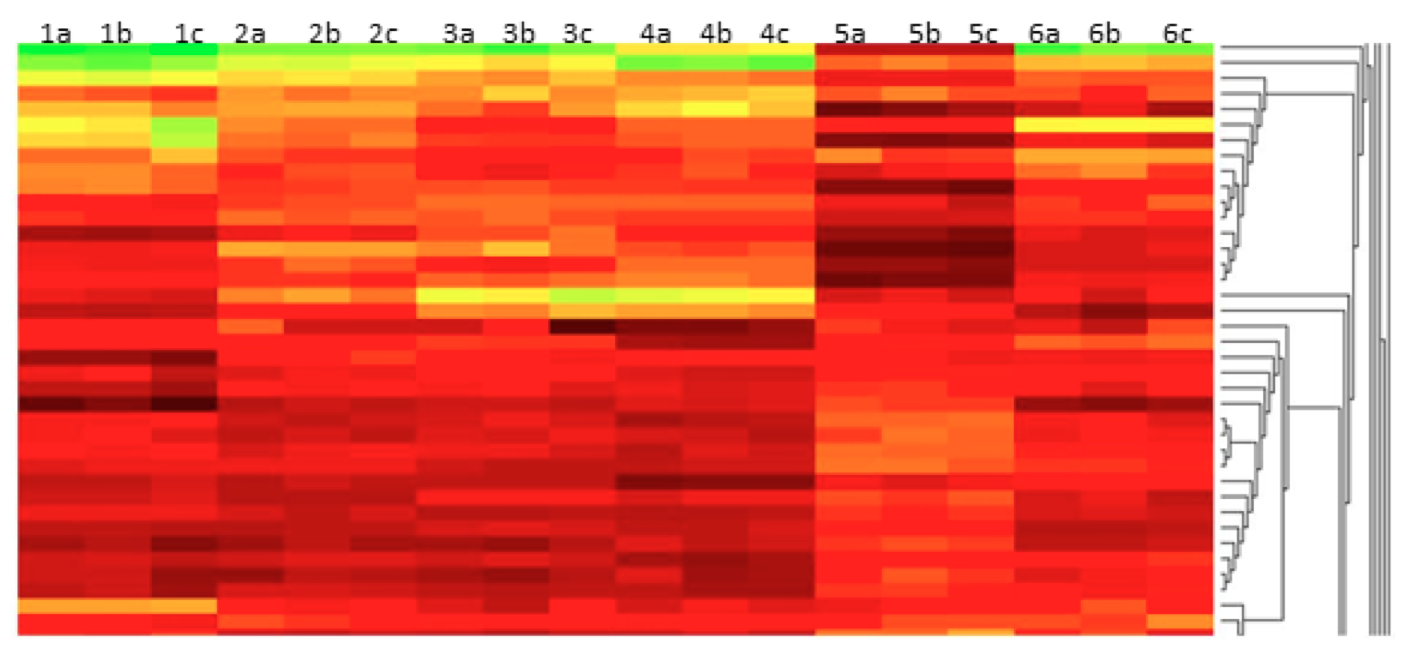
Figure 1: Whole transcriptome analysis of cells cultured under different growth conditions. Cell cultures were grown in triplicate under six different growth conditions. Gene expression analysis shows unique profiles consistent across triplicates for the six different conditions.
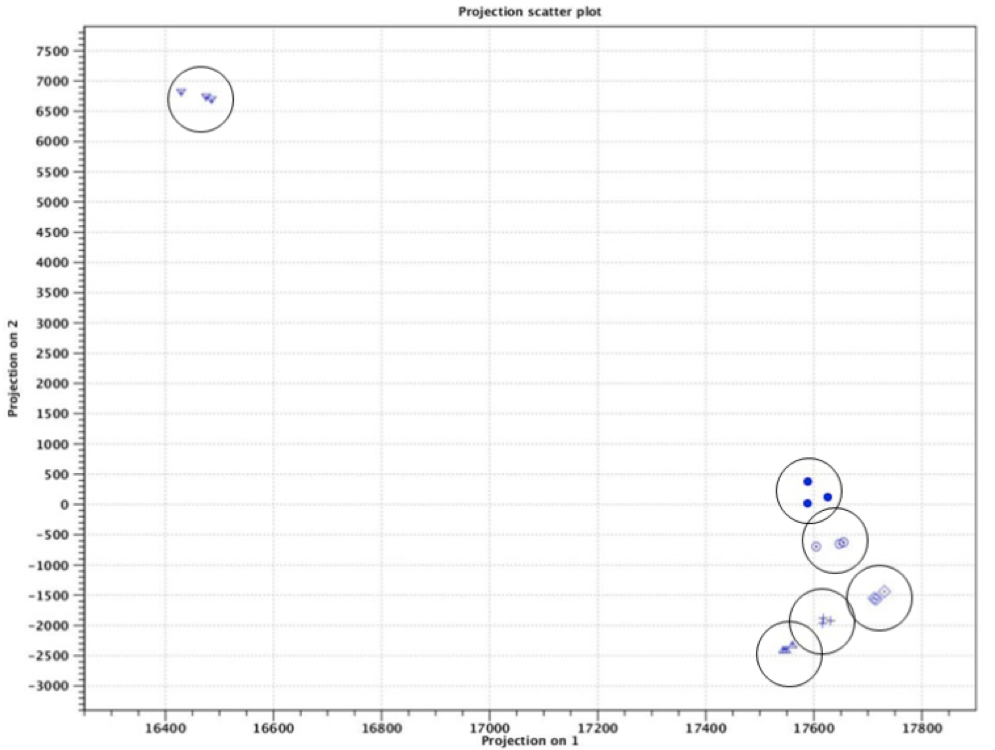
Figure 2. Principal component analysis clusters triplicate samples into 6 groups.
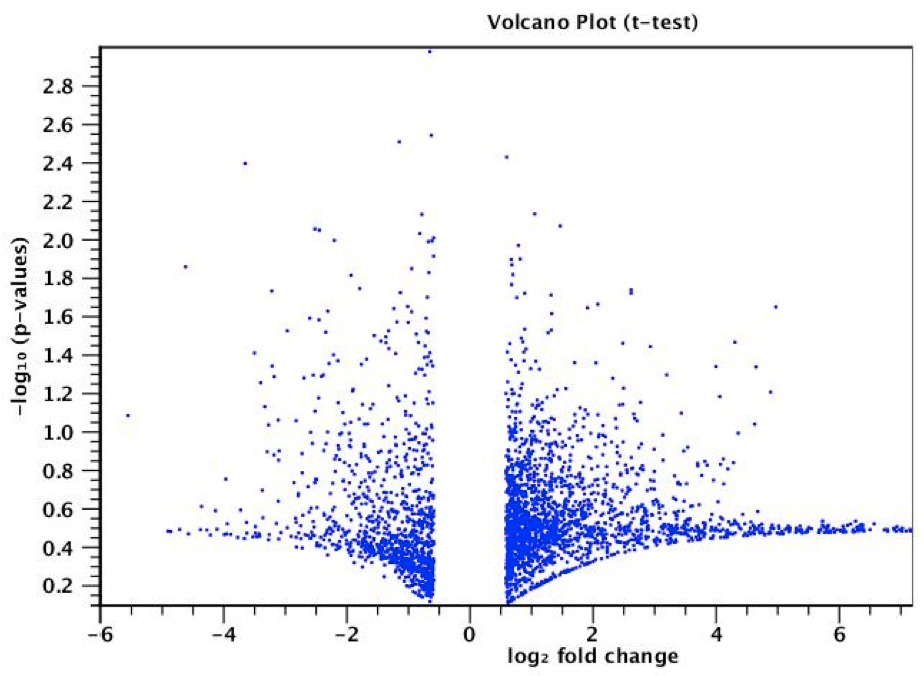
Figure 3. A volcano plot showing differential expression. It contains a point for each gene analyzed and plots log2(fold change) and –log10(p-value). Genes with significant expression changes (compared to a control) have a large magnitude fold change and high statistical significance (low p-value). These are the points towards the top of the plot.
Targeted RNA-seq
Targeted RNA capture allows you to isolate and analyze only your genes of interest, allowing for higher multiplexing of samples. The process consists of cDNA synthesis, followed by NGS library prep and target enrichment using probe-based capture. This approach is ideal for expression analysis of FFPE tumor specimens and can identify mutations, splice variants and fusions. Learn more about tumor genomic profiling here.
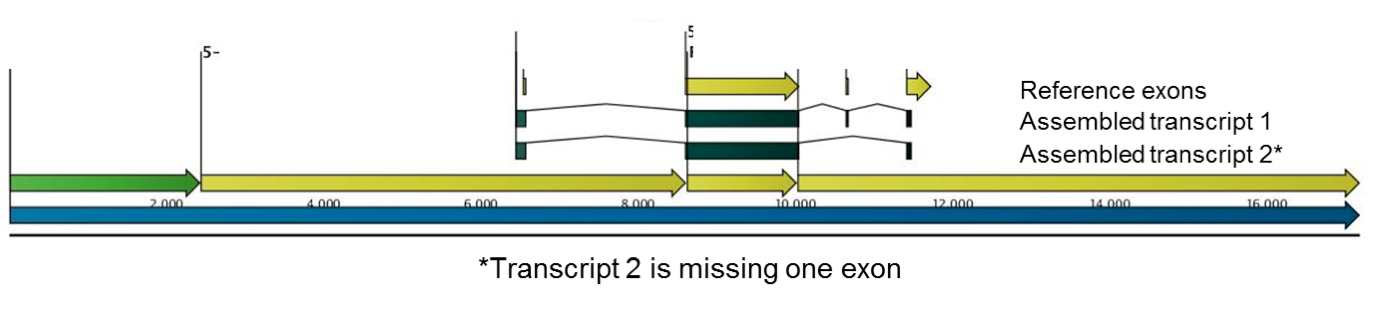
Figure 4. Targeted RNA-seq identifies a novel splice variant. Assembled transcript 2 is missing one exon when compared to reference.