Tumor Genomic Profiling
The development of targeted therapies for both germline and somatic DNA mutations has increased the need for molecular profiling assays to determine the mutational status of specific genes. Moreover, the potential of off-label prescription of targeted therapies favors classifying tumors based on DNA alterations rather than traditional tissue pathology. Ambry Genetics offers several platforms for tumor and tumor/germline paired analysis. Ambry Genetics has extensive experience designing and validating custom NGS assays and can provide this service for you.
What is TumorNext-142?
A custom NGS panel that can detect single nucleotide variants, small insertions and deletions in 142 genes that are frequently mutated in somatic and/or germline cancers [1]. TumorNext also detects gene fusions and structural variants, such as tandem duplications and inversions, in 15 frequently disrupted oncogenes and tumor suppressors. Through a matched control and custom bioinformatics pipeline this assays differentiates between somatic and germline mutations, allowing precise variant classification. Moreover, Jones et al. determined that tumor-only sequencing may result in a 31% - 65% false positive rate [2, 3], so incorporating a matched normal is critical for high efficient tumor profiling. See validation publication here.
Our tumor/normal assay and bioinformatics pipeline can be customized to meet your needs.
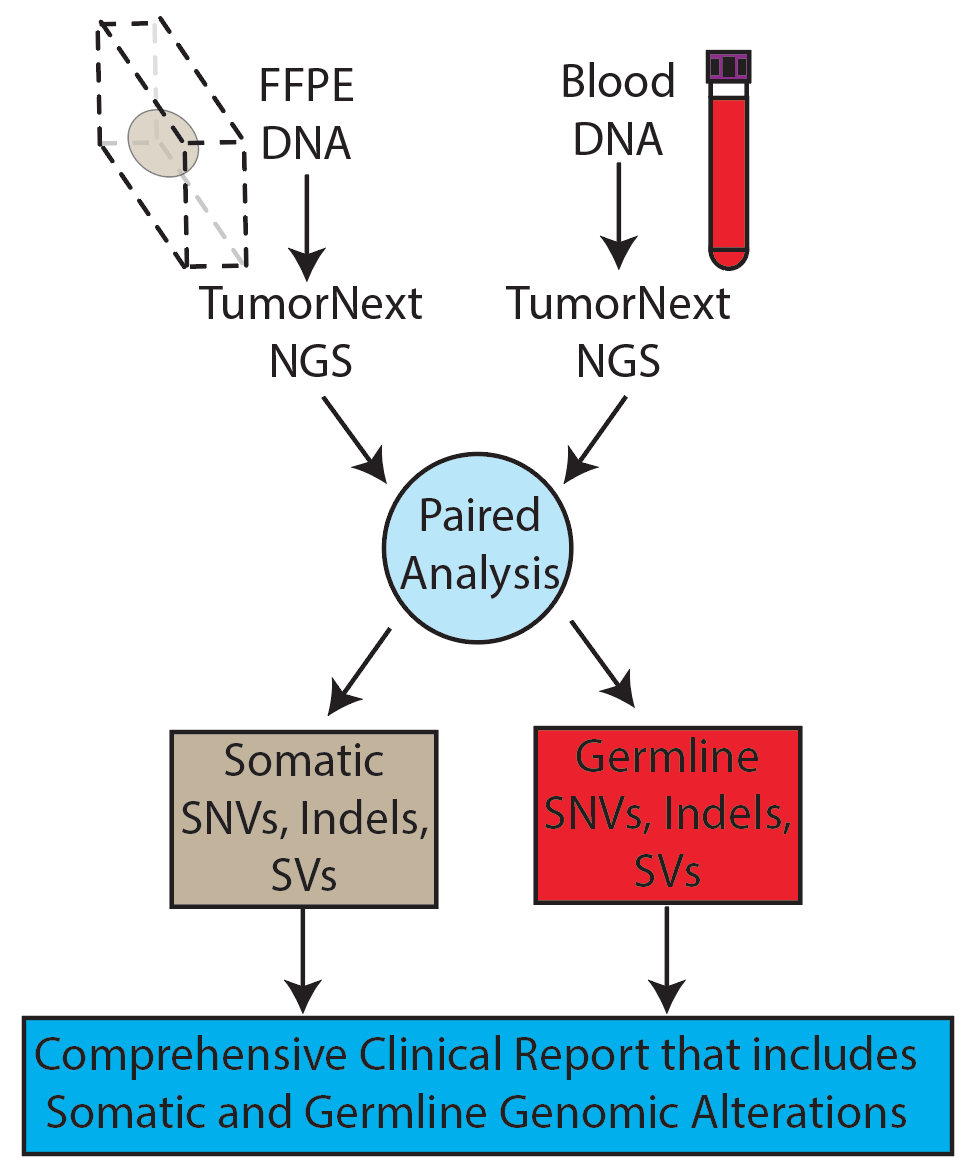
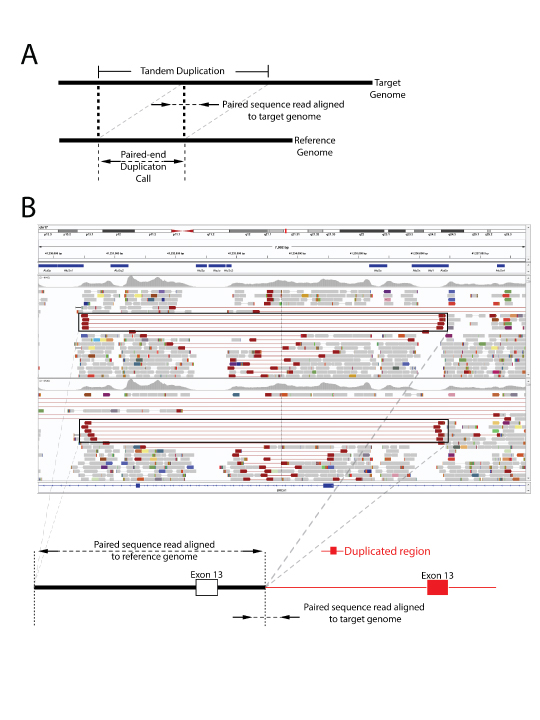
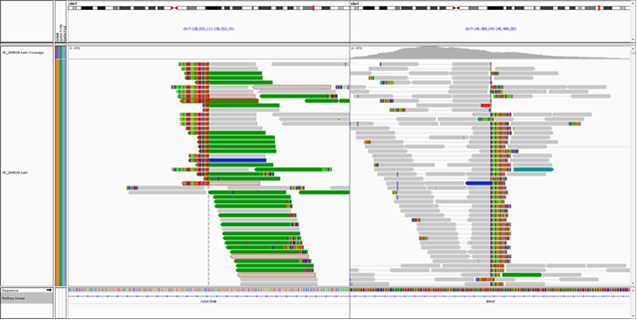
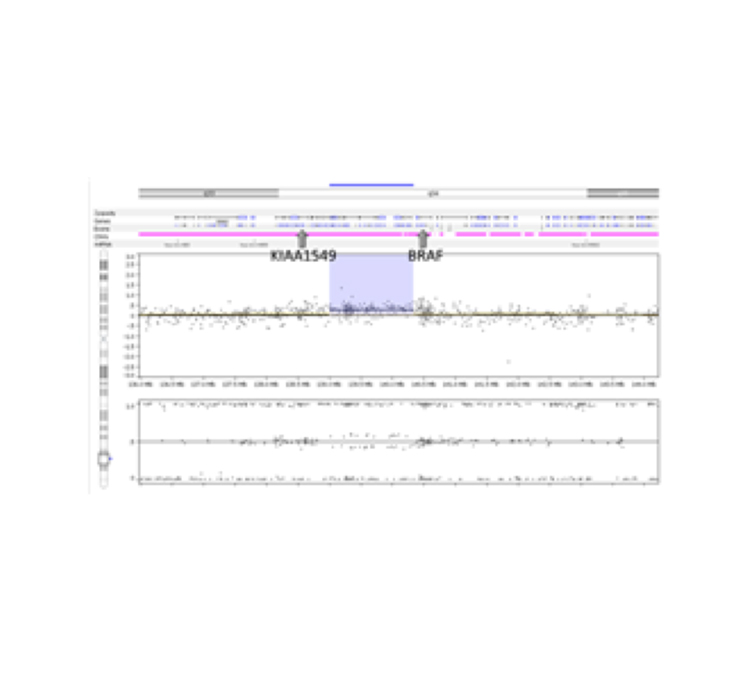
Left Panel: TumorNext workflow, Middle Panel: Tandem duplication detection by TumorNext. A) Schematic of tandem duplication detection using paired end reads. Reads are represented by arrows. Standard paired end reads face each other as represented in the target genome alignment. A tandem duplication will result in paired end reads facing away from each other when aligned to a reference genome. B) IGV screenshot of two tumor specimens with a known BRCA1 exon-13 6kb duplication and schematic of the duplication. The supporting paired end reads are located inside the boxes. Right Panel: Gene fusion detection by TumorNext. IGV screenshot of the KIAA1549-BRAF fusion showing high coverage of the breakpoint (Note: the multi-colored part of the read aligns to the fusion partner). This fusion is a result of a duplication in the intergenic region between KIAA1549 and BRAF, which was confirmed on the Affymetrix OncoScan array.
TumorNext-MMR-HRD
A focused NGS panel targeting genes in the mismatch and homologous recombination repair pathways. The assay can also provide microsatellite instability status and mutation burden.
| AKT1 | BRAF | CDKN2A | FANCA | MLH1 | PALB2 | RAD51C |
| APC | BRCA1 | CHEK1 | FANCD2 | MRE11A | PDGFRA | RAD51D |
| ARID1A | BRCA2 | CHEK2 | FANCI | MSH2 | PIK3CA | RAD54L |
| ATM | BRIP1 | CTNNA1 | FANCL | MSH6 | PMS2 | RPS20 |
| ATR | BUB1 | CTNNB1 | FBXW7 | MUTYH | POLD1 | SMAD4 |
| AXIN1 | BUB3 | DICER1 | GALNT12 | MYCN | POLE | SMARCA4 |
| AXIN2 | CCND1 | EMSY | GREM1 | NBN | PTEN | STK11 |
| BARD1 | CCNE1 | EPCAM | GRID1 | NF1 | RAD50 | TERT |
| BLM | CDH1 | ERBB2 | HDAC2 | NRAS | RAD51 | TP53 |
| BMPR1A | CDK12 | FAM175A | KRAS | NTHL1 | RAD51B | |
| Gold Standard MSI Markers: BAT25, BAT26, MONO-27, NR-21, NR-24 | ||||||
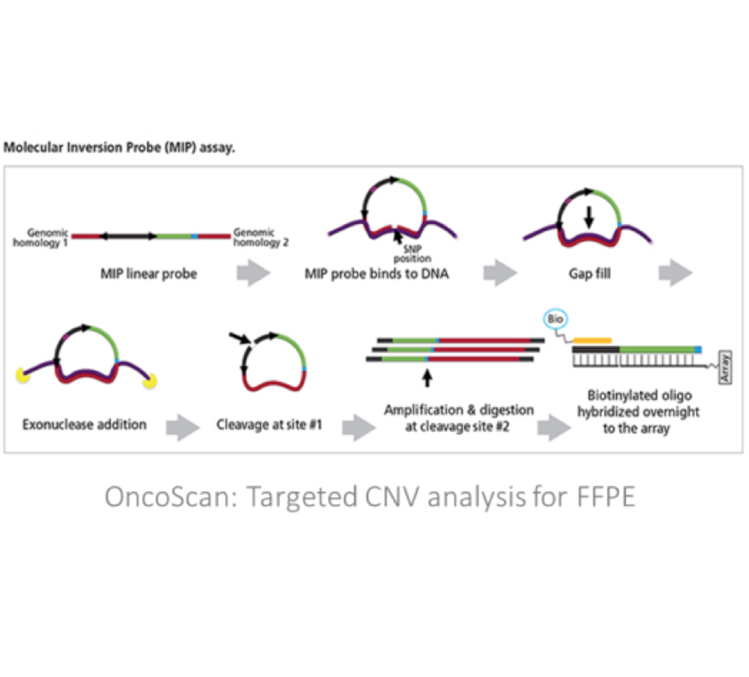
Affymetrix OncoScan
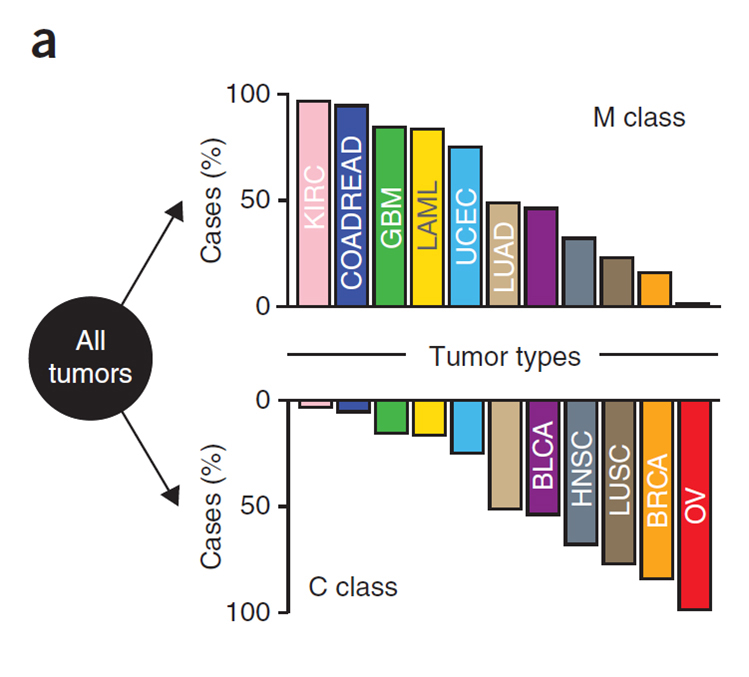
Based on Molecular Inversion Probe (MIP) target enrichment, the Affymetrix OncoScan array provides high resolution copy number variant detection, capable of detecting single copy amplifications, hemizygous deletions and copy neutral loss of heterozygousity (LOH). A recent paper by Ciriello et al. showed a bias in the type of mutations found in specific tumor types, such that specific tumors contained primarily either CNVs (C Class) or SNVs/indels (M Class), but not both [4]. Breast, ovarian, lung and head/neck cancers displayed the largest percentage of CNVs in the Ciriello study (see figure). As such, the OncoScan is an ideal platform when characterizing these tumor types.
- M class mutations - sequence alterations
- C class mutations - CNVs
- Tumors harbor predominantly CNVs or mutations, but not both
- Ciriello et al. Emerging landscape of oncogenic signatures across human cancers. 45(10) Nature Genetics, 2013
- Provides whole genome CNV analysis
- Can detect LOH: both hemizygous deletions and copy neutral LOH
- Hemizygous deletions are not detected using NGS methods for CNV analysis
- LOH is often the "second hit" in hereditary cancer patients
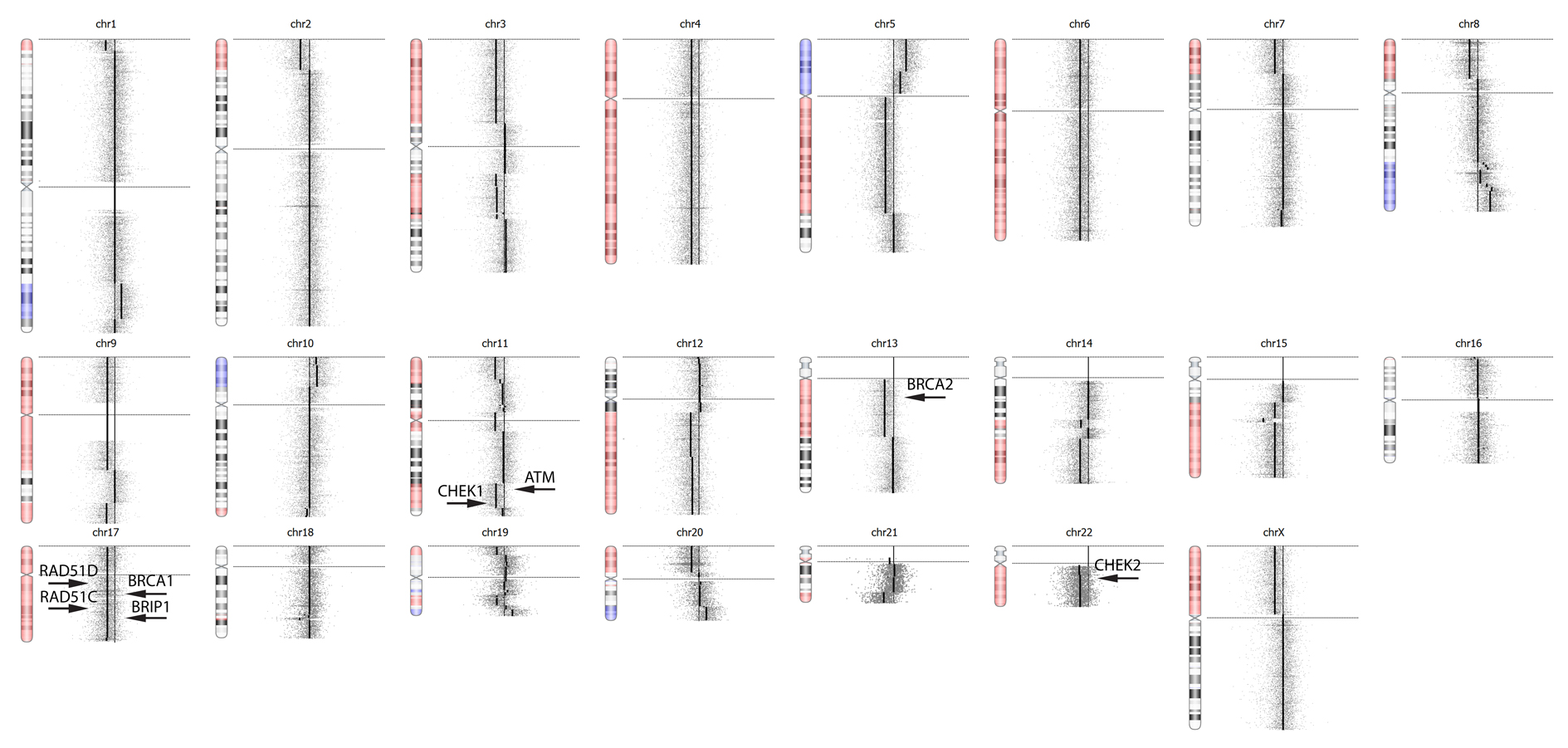
- Example karyotype: OncoScan shows hemizygous deletions in ATM, CHECK1, CHECK2, BRCA1, BRCA2, BRIP1, RAD51C, RAD51D, which represents 8 of the 13 genes involved in homologous recombination DNA repair.
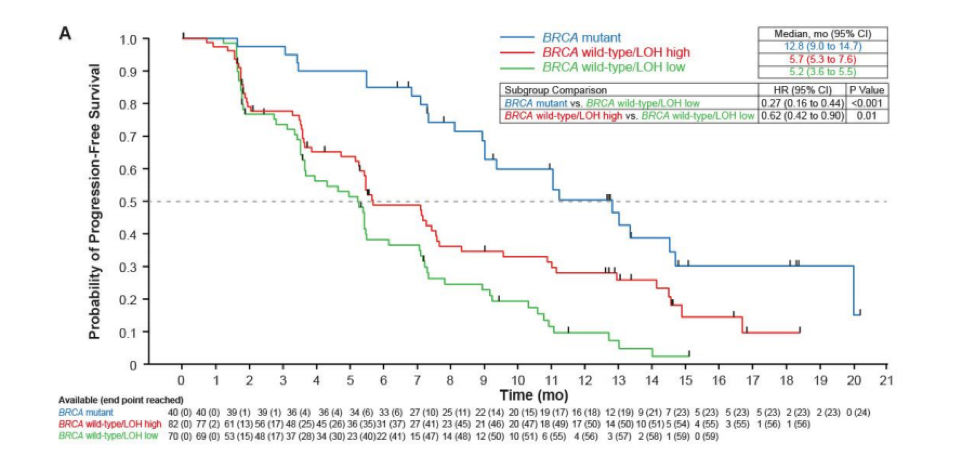
Loss of heterozygosity is a surrogate for homologous recombination deficiency (HRD). Several studies, including the PARP inhibitor trial Ariel2 [5], demonstrated that ovarian cancer patients with wild type BRCA1/2 alleles still responded to PARP inhibitors if greater than 16% of tumor cell genomes exhibited LOH. The Affymetrix OncoScan is a cost effective tool to determine global LOH profiles.
Final Efficacy Analysis Ariel2: PFS in BRCAmut and LOH-High Versus LOH-Low Patients
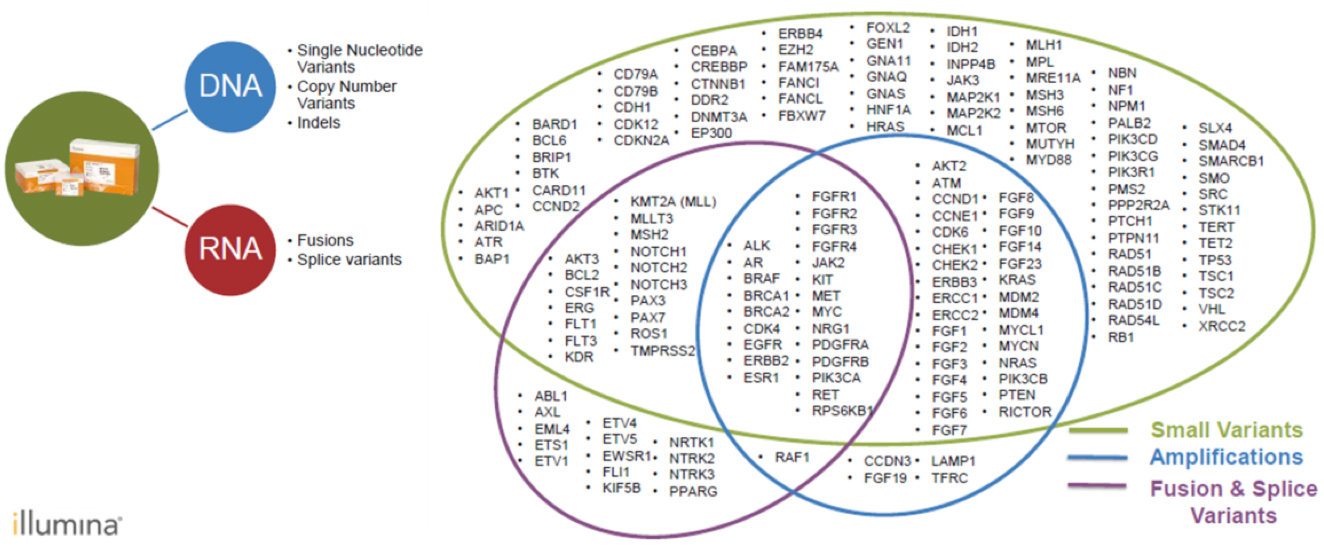
Illumina TruSight Tumor 170
TruSight Tumor 170, a next-generation sequencing assay designed to cover 170 genes associated with common solid tumors, is an enrichment-based targeted panel that simultaneously analyzes DNA and RNA, covering a wide range of genes and variant types. The comprehensive nature provides laboratories with a deep view into the genetics of cancer.
-
Comprehensive Coverage of Cancer-Related Variants
Assessment of fusions, splice variants, insertions/deletions, single-nucleotide variants (SNVs), tumor mutation burden and amplifications in one assay using DNA and RNA creates efficiencies in sample usage, time, and cost. -
Accurate Results from Low-Quality Samples1
Variant detection with as little as 40 ng DNA and RNA input, and as low as 5% mutant allele frequency, maximizes the results from precious formalin-fixed paraffin-embedded (FFPE) samples. -
Integrated, Streamlined Workflow
DNA and RNA are prepared in parallel with an integrated workflow following DNA shearing/cDNA synthesis.
TruSight Tumor 170 Gene List and Variant Classification
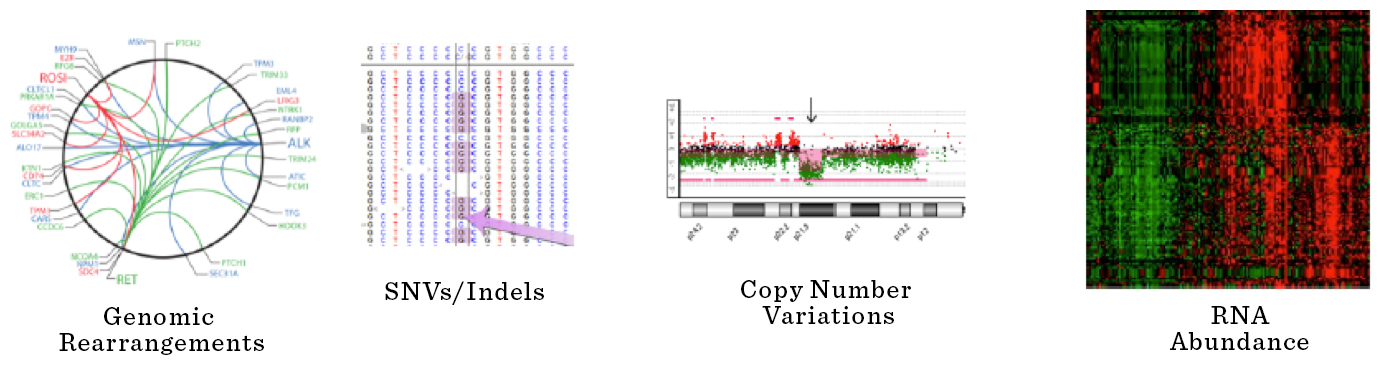
Archer Fusion and Variant Plex Assays
The Archer FusionPlex Assay system generates target-enriched cDNA libraries for NGS. The system leverages Anchored Multiplex PCR (AMP™) to selectively amplify cDNAs of specific genes of interest in a sample along with any fusion partners, known or unknown. Archer Analysis software can detect true positive calls for gene fusions (both known and unknown), isoforms, insertions/deletions (indels) and single nucleotide variants with as little as 20ng input nucleic acid from FFPE samples.
The Archer VariantPlex Assay system generates target-enriched DNA libraries for NGS using AMP™ technology. AMP enrichment is unrestrained by opposing primers, allowing larger genomic regions to be amplified and sequenced. AMP-enabled long-range enrichment can cover more relevant variants and strand specificity provides the flexibility to design primers that are optimized for coverage across challenging regions of the genome (e.g., GC-rich regions). Strand specificity also reduces allele dropout via dual independent coverage across target regions to ensure that some reads are retained when one primer drops out. AMP technology also leverages molecular barcodes to remove PCR duplicates, artifacts and sequencing errors, which enables robust, quantitative data that can be used for accurate variant calling and sensitive, concurrent CNV detection.
- Gray PN, Vuong H, Tsai P, Lu HM, Mu W, Hsuan V, Hoo J, Shah S, Uyeda L, Fox S, Patel H, Janicek M, Brown S, Dobrea L, Wagman L, Plimack E, et al. TumorNext: A comprehensive tumor profiling assay that incorporates high resolution copy number analysis and germline status to improve testing accuracy. Oncotarget. 2016.
- Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, Shukla M, Chesnick B, Kadan M, Papp E, Galens KG, Murphy D, Zhang T, Kann L, Sausen M, Angiuoli SV, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Science translational medicine. 2015; 7(283):283ra253.
- Karow J. (2015). Tumor-only Sequencing May Misguide Therapy But Many Labs Omit Matched Control. Genomeweb. (online: Genomeweb LLC).
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N and Sander C. Emerging landscape of oncogenic signatures across human cancers. Nature genetics. 2013; 45(10):1127-1133.
- Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, Konecny GE, Coleman RL, Tinker AV, O'Malley DM, Kristeleit RS, Ma L, Bell-McGuinn KM, Brenton JD, Cragun JM, Oaknin A, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. The Lancet. 2017; 18(1):75-87.